We use X-ray crystallography to obtain high-resolution atomic models of proteins of interest. The three-dimensional structure of a protein can contribute crucial insights into how a given macromolecule functions within a cell. A structural model provides an overall description of the protein as well as details about biologically significant regions of the molecule, interdomain interactions, binding sites for biological moieties important to the functioning of the protein as well as potential binding surfaces for cognate protein partners. Most importantly, such a structure could uncover a novel drug nucleus.
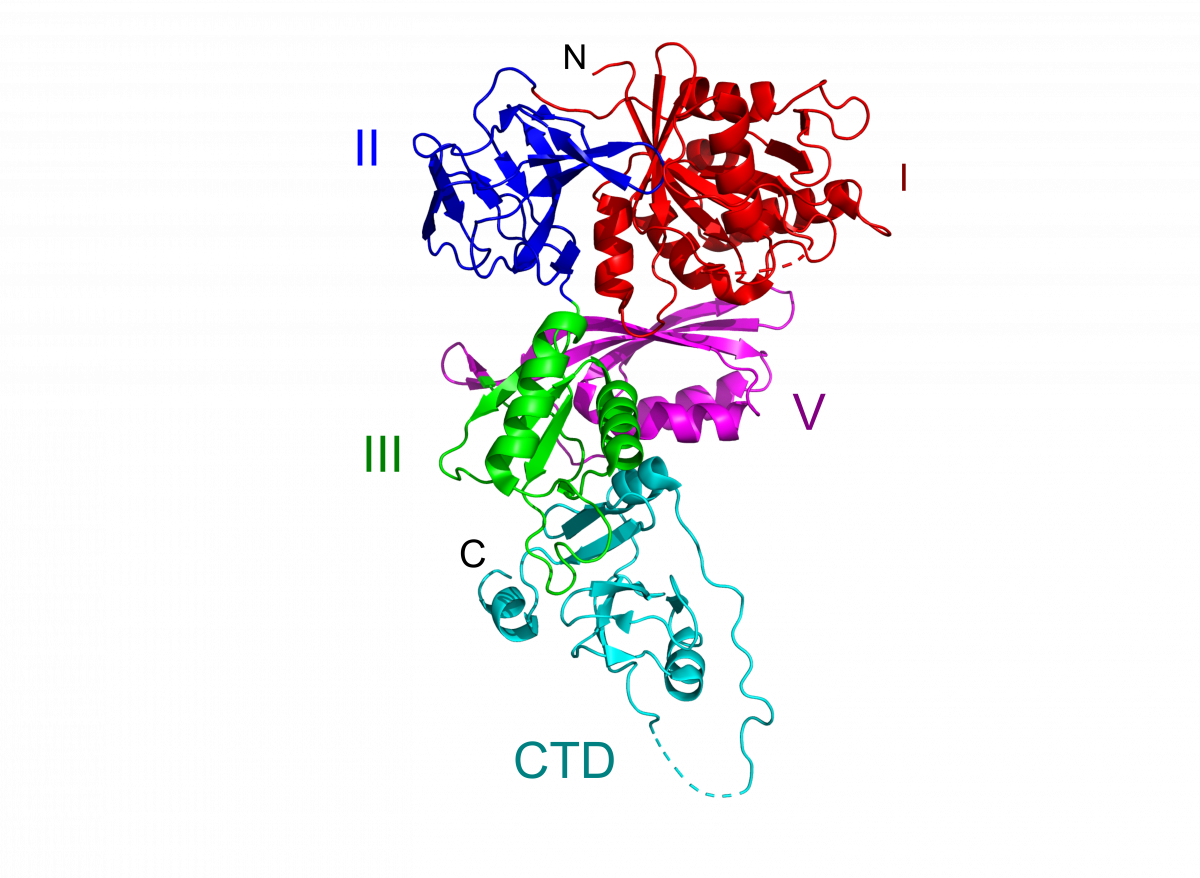
BipA
We have solved the crystal structure of Salmonella typhimurium BipA to 2.3 Å resolution by selenium phasing, with two copies of the protein in the asymmetric unit. In agreement with sequence homology, the first four domains in BipA resemble domains I, II, III and V in the EF-G/EF-2 family of translation factors. The C-terminal domain of BipA, which is positioned similarly to domain IV in EF-G, has a unique fold. Although the overall topology of these domains is similar in the two proteins, it is readily apparent that their relative orientations within the two structures are unique. (PDB 8EWH)
Brown, R.S. et al (2007) Crystallographic and Biochemical Characterization of the GTPase and Ribosome Binding Properties of Salmonella typhimurium BipA. J Biomol Struct Dyn, 24:6: 630-630.
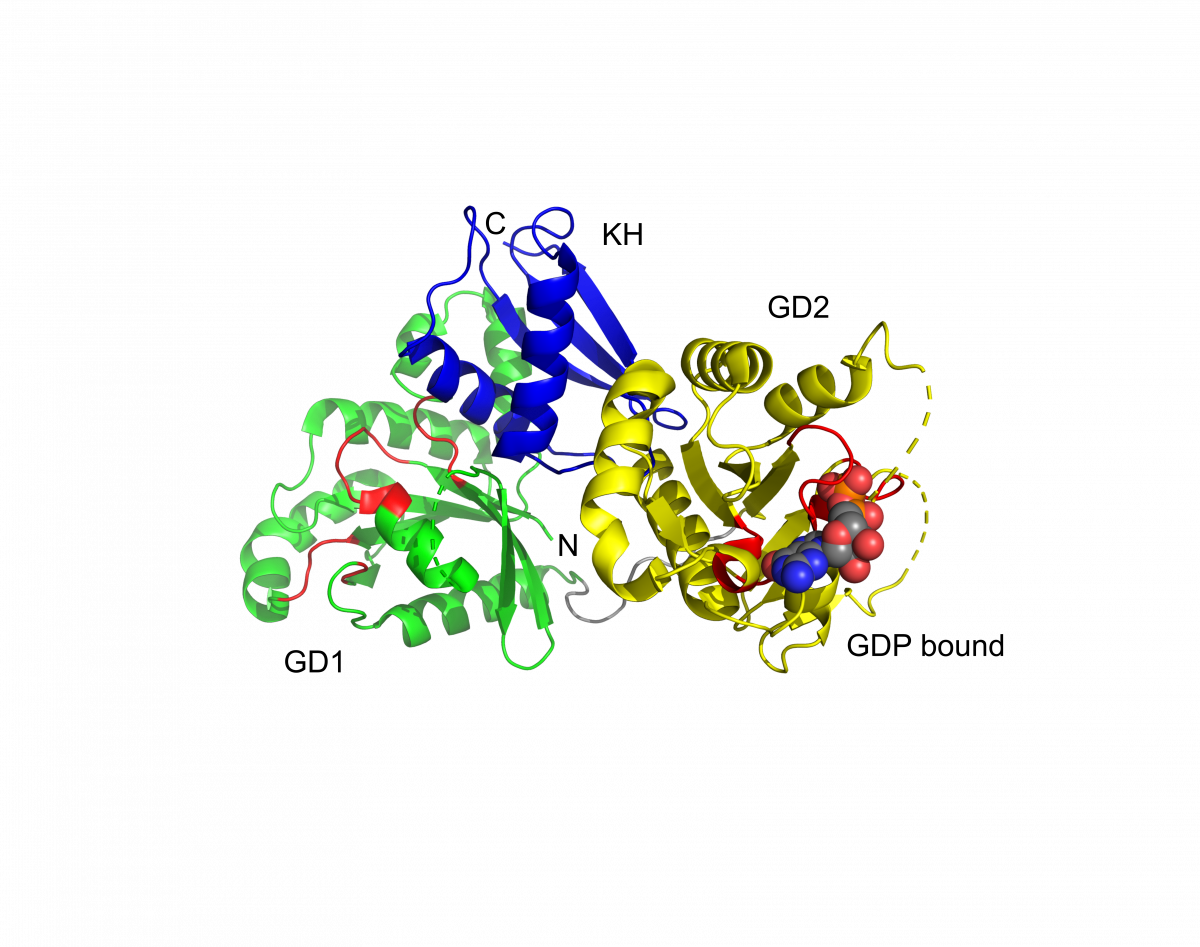
EngA
Ribbon diagram depicting the overall fold of the EngA protein, with each of the three domains in a different color. GTPase domains 1 and 2 (GD1 and GD2) and the C-terminal (KH) domain are colored green, yellow and blue, respectively. Residues corresponding to the conserved motifs of the nucleotide binding site in each GTPase domain are colored red. The GDP bound to the second GTPase domain is shown as a space-filling model. Undefined regions are indicated by dashed lines and correspond to the switch I region of GD1 and the switch I and II regions of GD2. (PDB 1MKY)
Robinson, V.L. et al (2002) Domain arrangement of Der, a switch protein containing two GTPase domains. Structure, 10: 1649-58.